Towards EV mimics
As a first possible strategy, we are now implementing selected mixtures of artificial lipids creating a “tailor made” vesicles and adapting it to efficiently encapsulate the candidate porous nanoparticles. As the final artificial EV should behave as the natural one, this artificial structure can be decorated with additional targeting or functional moieties, such as peptides, proteins/antibody for the cell target recognition, fluorescent dyes for ease optical tracking, PEGylation for effective extension of the nanovesicles half-life in the blood stream, charged lipids able to boost up intracellular uptake in the target recipient cells. This procedure guarantees also an enhanced colloidal stability of the encapsulated nanoparticle and the increase in the cargo encapsulation efficiency thanks to the use of a porous vehicle.
Therefore, the complexity of the synthetic system will always be inferior to the biological counterpart, which is both a strength, since the system counts less variables and is easier to manipulate, but also the major drawback, since a necessary (and difficult) simplification is implied in the identification of the key components of the natural elements. The risk is that simplification could be too gross to reach the same performances in terms of the delivery efficacy of natural EVs. This type of approach is, in general, very challenging, because it requires a detailed understanding of the role of many of the elements composing natural vesicles and the ability to combine them.
Artificial EVs represent the last frontier toward which research is heading, in trying to understand how to solve all the aforementioned limitations. Despite the promising results presented in vitro, this field is still in its infancy and it will take time to bring them to in vivo studies and possibly clinical applications.
EVs derived from tumor/osteoblastic cells
To identify molecules on natural EVs that can mediate targeting to bone cells, we selected the PC3 and PC3M cell lines. The PC3 cell line originated from a bone metastasis of a prostate tumor (human). PC3-derived EVs have been reported to interact with bone cells, including osteoclasts (Tamura et al, Int J of Mol Sci, 2020). PC3M is a derivative cell line with higher metastasizing potential.
Following our conventional protocol for EV isolation from cell culture supernatant, we first enriched for EVs by differential (ultra)centrifugation and further purified EVs by density gradient ultracentrifugation. We verified the presence of EVs using high-resolution flow cytometry and western blot analysis. Our data indicate that EVs from PC3 cells have buoyant densities of 1.04- 1.10 g/ml, and contain the characteristic tetraspanin proteins CD9, CD63. We also analyzed the proteomes of purified PC3 EVs and source cells by MS/MS. As much as 3600 unique proteins were detected in EVs and many of these were enriched compared to the source cell proteome. The top EV proteins detected were actin, integrins (including β4, β1, α2, α3) and CD44, heat shock proteins (90, 70, 27), and the tetraspanin proteins CD9, CD81 and CD63. Together, these data indicate that PC3 EVs can be purified and collected in sufficient quantities to allow in-depth proteome analysis.
Molecular models and simulations
At this stage of the project, we have set up different models of the lipid bilayers that could be adopted to constitute the envelopes of artificial extracellular vesicles (EV). We are usin g these models to characterize their
physical properties via molecular dynamics simulations (MD) and gain insights on what lipid bilayer compositions could be more suitable for the functional artificial EV. The studied membrane compositions have been propo
sed by experimental partners (PoliTo/IRFMN groups) as relevant candidates to model naturally observable EV (Ferreri et al, Cancers , 12 (4), 900, 2020), while retaining a relatively simple composition. Indeed, for effective
molecular modeling, these composi tions have been slightly simplified from the typical natural EV compositions by eliminating lipid types that represented less than 1% of the system. To describe the behavior of these
systems in MD simulations, we employ the well established Martini force field (Souza et al, Nat. Methods, 18, 382 388, 2021). This coarse graining (CG) approach describes molecular fragments and chemical groups as
interacting CG spheres, which allows to simplify the representation of molecular systems (3 4 heavy atoms per CG s phere) while preserving their important features. This enables the simulation of space and time scales relevant
to the study of complex systems such as our putative EV mimics envelopes. It is important here to underline the fact that only CG modeling sche mes allow studying the large scale dynamics of such systems. Higher
resolution models (all atom) cannot be applied in this case, because of the large size of the molecular systems and the long simulation timescales necessary to observe dynamic events and reorganization within the EV envelopes. Our
choice of the modeling scheme follows our recent work on optimizing Martini CG lipid models to better match both experimental measurements and the behavior of atomistic model simulations (Empereur Mot et al, J C hem Phys,
156, 024801, 2022). We here developed an automatic optimization strategy which was employed to calibrate and optimize the parameters of the existing, state of the art Martini CG lipid models. Although improved agreement
with experiments was obtai ned in some cases (notably, optimized models reproduce the correct phase behavior of lipid mixtures at low temperatures, Fig. 1, which is poorly described using current Martini models) more work
is needed in order to obtain the next production version of t his force field that will be usable for our simulations of EV mimic envelopes. In particular, the current Martini Cholesterol model also needs to be re parameterized
for compatibility with the new lipid models. In the meanwhile Martini 2.0 includes the bes t validated Cholesterol model available to date (Melo et al, JCP , 143, 243152, 2015), as well as compatible parameters for PEG
chains (useful for modeling PEGylated DSPE lipids) and lipid models accurate enough for our modeling needs of the EV mimic envelopes. In order to compare properties of different EV mimic envelopes, assessing the impact
of the composition on the structure and dynamics of the membranes, we have first simulated systems representing the selected compositions of MIMIC 1, MIMIC 2 and MIM IC 3 as free bilayers in solvent, as well as
the three same envelope compositions with a cholesterol content increased by 30% (Fig. 2A). Bilayers were made of 1000 molecules each and simulations were run at a timestep of 20 fs for 10 µs at P=1 Atm and T=31 0
K, after an equilibration stage of 1 µs.
In these simulations, we are primarily interested in measuring the lateral diffusion of the lipid heads, which provide a measure of the mobility of the lipids across the surface of the membrane (Fig. 2B). This assessment of the lipid
dynamics is of primary interest in the choice of the lipid composition to be used for constituting the EV mimic envelope. These results highlight the impact of the Cholesterol content on the lateral compressibility and diffusion
coefficient of the membrane lipids (Fig. 2C). To this end we measured the area compressibility modulus (KA) in Fig. 2C, which corresponds to the elastic energy produced by the stretching or compression of a bilayer patch with respect
to its equilibrium area, as well as other structural properties such as the thickness and area per lipid, assessing the variations determined by the choice of lipid composition
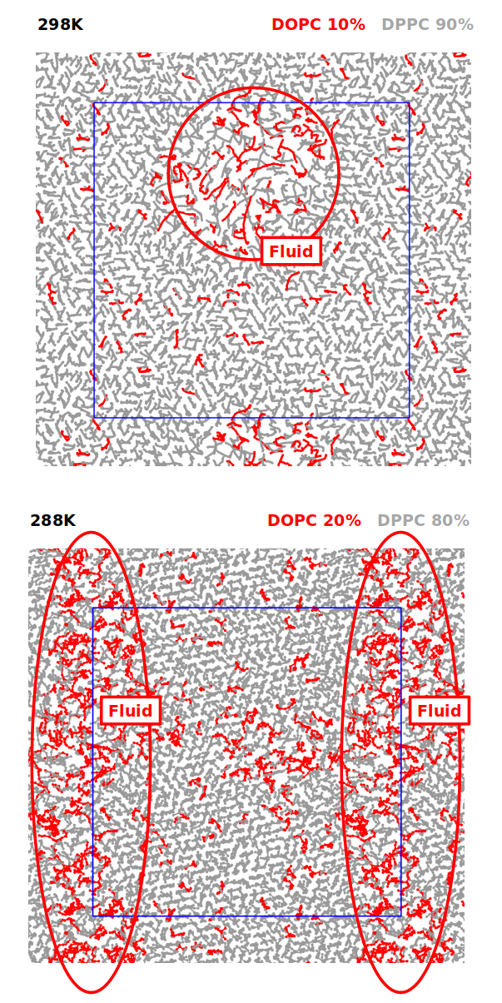
Fig. 1: (top) Simulation of a bilayer composed of DOPC and DPPC at 10 and 90% mass, respectively (orthogonal view). The system clearly exhibits areas of fluid and gel phases, with different packing and diffusivity of the lipids. (bottom) Simulation of a bilayer composed of DOPC and DPPC at 20% and 80% mass, respectively (orthogonal view). The system exhibits a larger area of lipids being in the fluid phase, due to the increased percentage of DOPC lipids.
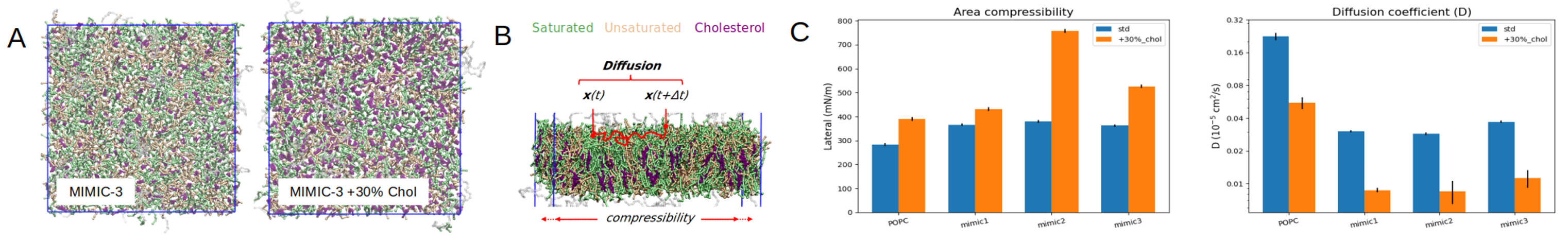
Fig 2. (A) Orthogonal view of the MIMIC-3 bilayer with or without 30% cholesterol increase. (B) Illustration of the principle of computation of the diffusion coefficient of the lipids within the upper leaflet of the bilayers. (C) Area compressibility modulus (K A) and diffusion coefficient calculated over 10 us of equilibrium simulation for MIMIC-1, MIMIC-2 and MIMIC-3, with or without 30% cholesterol increase. A pure POPC bilayer is also included for comparison.
We then proceeded to model the grafting of membrane envelopes onto the surface of solid organo-silica particles. Starting from the simulation results of the free bilayers at different compositions we selected the model that exhibited
the largest lateral diffusivity, i.e. the composition that guarantees the best intramembrane mobility, namely the MIMIC-3 membrane. To mimic the impact that grafting can have on such supramolecular architectures, we first adopted
a qualitative modeling scheme and proceeded to restrain the position of the lipid heads of the lower leaflet of the bilayer (Fig. 3A). To this end, we applied external harmonic potentials to create position restraints on the relevant
phosphate CG spheres and assessed the diffusivity of the non-grafted upper leaflet lipids, to be compared with that of the free MIMIC-3 bilayer as a control case (Fig. 3B). Proceeding in this way could allow to systematically explore
the impact of membrane grafting on the mobility of the lipids within the supramolecular structure. For MIMIC-3 results show that lateral diffusion is reduced by ~20% in the upper leaflet, when the lower leaflet is grafted (Fig. 3B).
Having validated that such simulation setups with grafting exhibit notable and quantifiable differences in the upper leaflet dynamics, we then proceeded to improve our setup by explicitly representing the surface of hard-shell nanoparticle
that constitute the core of EV mimics. To this end we developed a model of organo-silica surface, to be introduced as solid substrate of the lipid bilayers in our MD simulations (instead of simply using position restraints on the
lipid heads). We also introduced edges to the geometry of the “hard” surface (Fig. 4), to mimic the effect of nanoparticle edges. To this purpose we are currently experimenting with membranes having two edges (one convex on the upper
leaflet side and the other convex on the lower leaflet side, as required to maintain periodic boundary conditions), with opening angles of 165° and 150° (with respect to the standard flat membrane with opening angle of 180°). We
then expect to be able to observe local re-organizations of the envelopes, with modified membrane compositions at the locations of the edges. The automated protocol we have created to this end is easily adaptable to modify the value
of the edge angle, or even to create edges of different shapes, according to the size and shape of the organo-silica nano-particle eventually selected by the experimental partners.
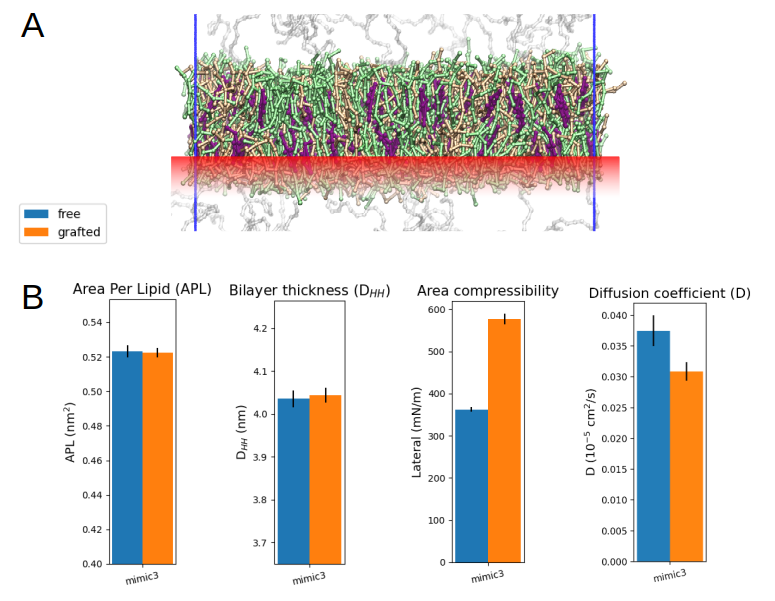
Fig 3. (A) Illustration of the harmonic position restraints (red) applied to simulate the grafting of the lower leaflet of a lipid bilayer, using MIMIC-3. (B) Structural and dynamic characteristics of the grafted versus non-grafted MIMIC-3 bilayer.
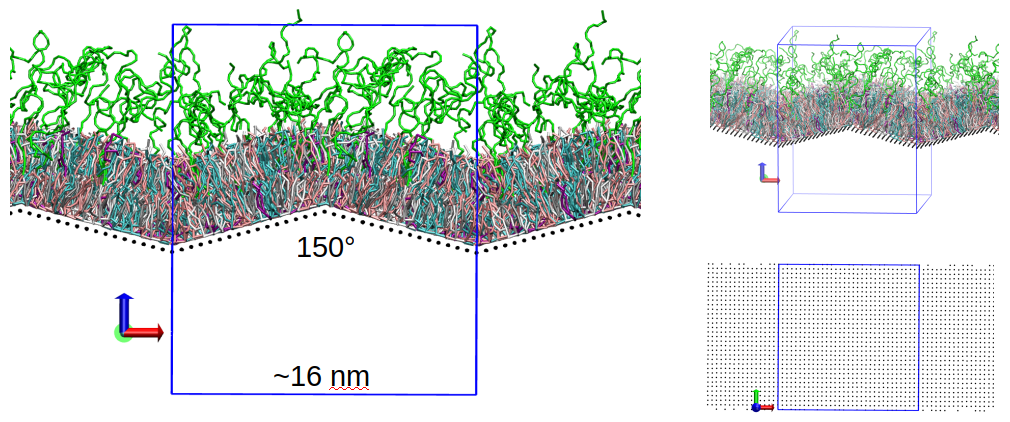
Fig 4. (left) Snapshot of a simulation box including MIMIC-3 and a preliminary version of the “hard” surface with edge angles of 150°. (top right) Another view of the same system. (bottom right) Orthogonal view of the lattice used to represent the “hard” surface.
Lastly, we are refining the explicit representation of the organo-silica surface to include its surface functional groups, starting with the diaza-silane functionalization (DMDASCP). We have started with creating an homogeneous single-layer grid of CG spheres representing the organo-silica surface structure (Fig. 4), based on the surface density descriptions available in the litterature (Emami et al, Chem. Mater, 26 (8), 2647-2658, 2014). We have then created additional CG spheres representing the DMDASCP groups (Fig. 5B), using simulation parameters compatible with the Martini 2.0 description of our systems that could be adapted from the literature (Pérez-Sánchez et al, Chem. Mater, 28 (8), 2715-2727, 2016). These serve as first intention to demonstrate the relevance of our modelization and remain to be further validated based on experimental evidences provided from the experimental partners. Interestingly, we noticed that the adsorption of bilayers onto the porous organo-silica surfaces might trigger a relocation of a meaningful amount of the cholesterol content of the membrane into its upper leaflet (Fig. 5B). Although this is only preliminary observations, we expect that more realistic representations of the organo-silica surface, its functionalization, size and shape, will provide valuable insights on large-scale membrane re-organization events such as this putative cholesterol relocation in between leaflets.
Data-driven analysis techniques (Gardin, et al, Commun. Chem., 5, 82, 2022) will be applied once we finish validating our modeling procedures, in order to investigate the local structure and dynamics of different molecular domains within the envelopes. Such local and dynamic-based characterization of the collective lipid behavior will provide insights on the intra-membrane dynamics, assisting the design of optimal EV mimics.
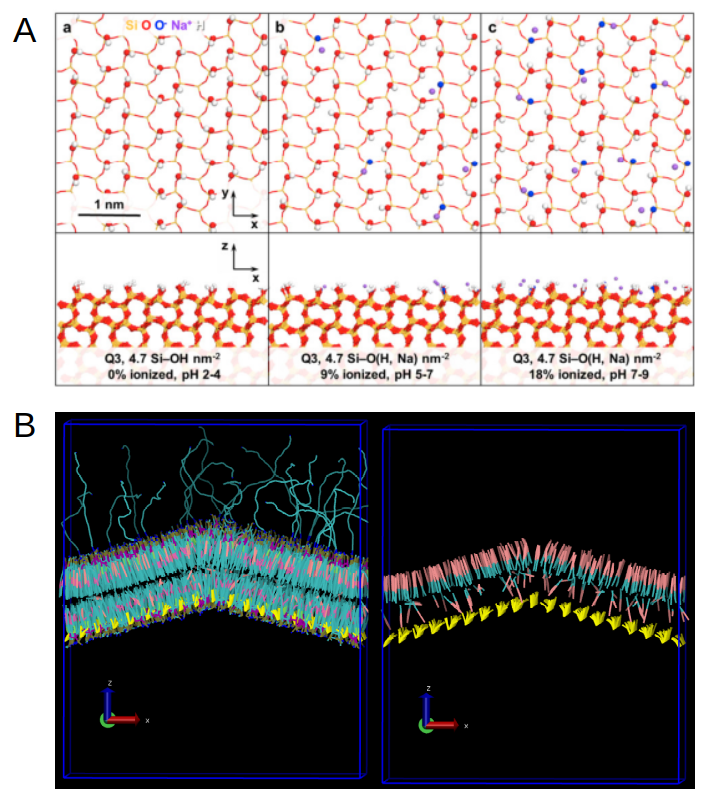
Fig 5. (A) Different ionization states of the organo-silica surfaces as described from atomistic simulations (Emami et al, Chem. Mater, 26 (8), 2647-2658, 2014). (B) Left: Side view of the MIMIC-3 bilayer grafted to the DMDASCP-functionalized organo-silica surface, shown in yellow. Right: Side view of the same bilayer with only cholesterol displayed in pink and cyan, showing an increased amount of cholesterol relocated to the upper leaflet of the envelope.
Public available results and resources
Patent applications
Italian Patent Application N. IT102023000015396 of 21th July 2023 “Nanoparticelle rivestite e loro uso per il trasporto di agenti terapeutici e diagnostici”, Inventors: V. Cauda, G. Rosso, M. Conte, L. DeCola, M. Sancho Albero, pending evaluation.

